Research
Cancer Genomic instability
-
Genomic instability as a major driving force of tumorigenesis
-
Cancer development is a complex multi-step process, involving a wide network of cellular events. Genomic instability, a hallmark of cancer, enables the acquirement of new characteristics required for tumorigenesis. We focus on the different cellular pathways which are altered under aberrant oncogene expression, causing various cellular stress reactions leading to perturbed DNA replication dynamics and chromosomal instability. We have shown that aberrant oncogene over-expression forces DNA replication to proceed under suboptimal conditions such as nucleotide deficiency, leading to replication stress and DNA damage (Bester et al. Cell 2011). Recently, we have demonstrated that activation of the RAS oncogene perturbs DNA replication due to increased topoisomerase 1 expression, disturbing the regulation of R-loop homeostasis and resulting in genomic instability (Sarni et al. Cell Reports 2022).
Another aspect of our work focuses on the tumor suppressor gene TP53, known as the “guardian of the genome” as it plays a key role in restricting tumor initiation and progression. We study the mechanisms underlying genomic instability including the catastrophic chromothripsis instability event, associated with aggressive tumorigenesis in cells deficient for wild type p53. We found that p53 loss leads to replication stress resulting in genomic instability. We found that nucleotide supplementation rescues replication stress, DNA damage and proliferation crisis (in progress).
Altogether, unfolding the molecular basis underling genome instability in cancer may open new potential prevention and therapeutic approaches.
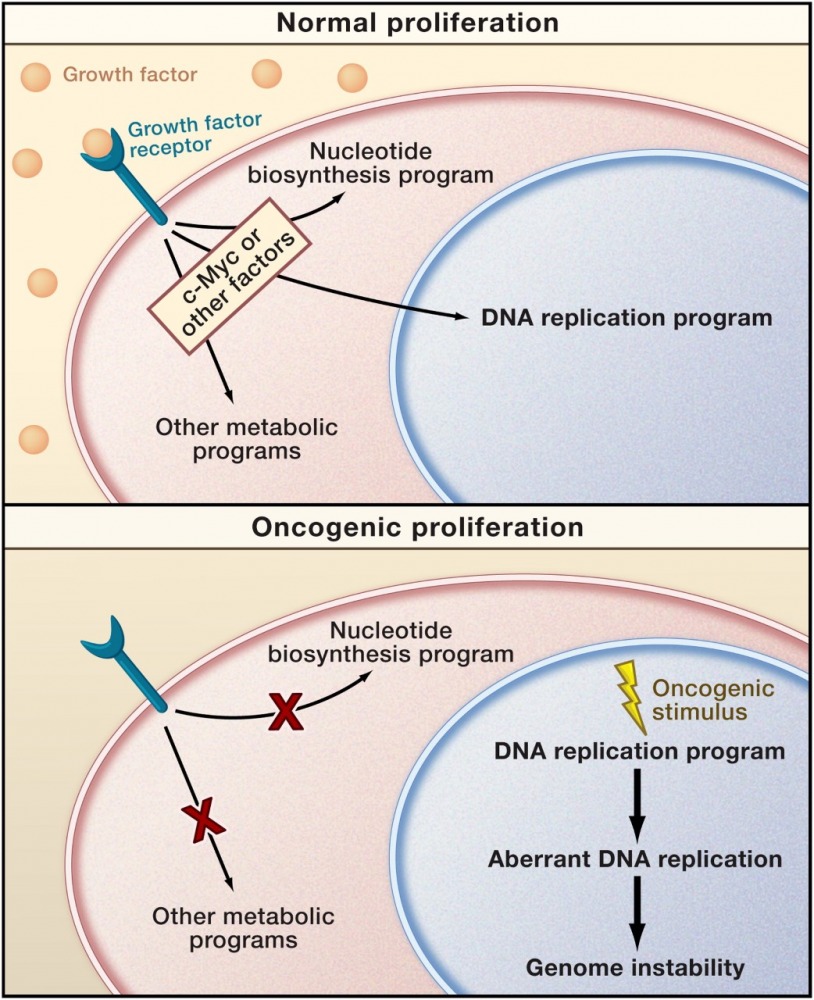
A mismatch between proliferation and metabolite production may characterize oncogenic cell cycles (Venkitaraman 2011, Preview in Cell on Bester et al., Cell 2011).
-
Recurrent chromosomal breakage and cancer
-
Common fragile sites (CFSs) are genomic regions susceptible to replication stress and are hotspots for chromosomal instability in cancer. CFSs instability accelerates tumorigenesis by increasing the probability of acquiring genomic alterations, supporting cancer progression. We aim to understand the features underlying their sensitivity during transformation.
We have previously shown that the repertoire of fragile sites is dynamic and dependent on the replication stress inducer. We demonstrated that the replication inhibitor Aphidicolin and various oncogenes lead to a different landscape of recurrent fragility, reflecting the variability in genomic alteration among tumors overexpressing different oncogenes (Miron et al. Nature Communications 2015).
Recently, we explored the transcriptional profile, DNA replication timing and the 3D genome organization under aphidicolin treatment and overexpression of various oncogenes. Analyzing the aphidicolin-induced fragility revealed a fragility signature, comprised of a topologically associated domain (TAD) boundary overlapping a highly transcribed large gene with replication timing delay. CFS stability may be compromised by incomplete DNA replication and repair in TAD boundaries core fragility regions leading to genomic instability (Sarni et al. Nature Communication 2020).
Currently, we are studying the mechanisms underlying oncogene induced fragility. Our preliminary results indicate the role of transcription perturbation in fragility (on-going projects). Our goal is to establish a strategy that can predict which pathways are disrupted in a given cancer type according to its chromosomal breakpoint pattern.
Altogether, identifying the various fragility signatures will allow a more comprehensive understanding of the role of CFSs in cancer development and will highlight the various mechanisms promoting genomic instability in cancer.

Delayed replication of highly transcribed large genes in TAD boundaries leads to DNA breaks.
Personalized medicine for Cystic Fibrosis patients carrying rare CFTR mutations
- Rare Mutations
-
Cystic Fibrosis (CF) is the most prevalent lethal genetic disease in Caucasians caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene. The triple combination of modulators (TRIKAFTA) was approved for ~80% of CF patients. Still, many patients carrying rare mutations are not eligible to this treatment, due to limited accessibility to patients’ samples and lack of clinical trials. The use of ex-vivo models, such as rectal organoids, enables testing of CFTR modulators in these individuals, predicting the possibility of a clinical benefit. Spatial CFTR structures, modeling the binding of the different TRIKAFTA components, shed light on the functional effects of the modulators on the mutated CFTR. These structural predictions enable the identification of candidate rare mutations that may benefit from the treatment. Our current aim is to use this new data to prioritize the assessment of the response of patients carrying rare mutations to TRIKAFTA and further shed light on the basis of the CFTR response to various modulators.
- Splicing Mutations
-
Cystic Fibrosis (CF) is the most prevalent lethal genetic disease in Caucasians caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene. We are studying different molecular mechanisms acting as modifiers of disease severity and response to therapy.
Our goal is to develop personalized medicine for CF patients carrying rare CFTR mutations including splicing and nonsense mutations (see below). For this, we are collecting patient-derived respiratory and intestinal epithelial cells that recapitulate essential features of the in vivo epithelium and exhibit CFTR-dependent functional readout. These patient-derived cellular models serve as a pre-clinical tool in the development of personalized therapies.
A significant fraction of CF-causing mutations (10-15%) affects pre-mRNA splicing, leading to the generation of aberrantly spliced CFTR transcripts. We are now developing a targeted modulation approach aimed to correct the splicing pattern of CFTR transcripts, according to the specific mutation. This strategy, which is based on splice-switching Antisense Oligonucleotides (ASO) is aimed to inhibit or activate specific splicing events by a steric blockade of the recognition of specific splicing elements. Together with the drug development company SpliSense, we have demonstrated the potential of this strategy in human respiratory cells (HNEs, HBEs) derived from CF patients carrying the 3849+10kb C-to-T splicing mutation. A highly potent ASO was able to significantly increase the level of correctly spliced CFTR mRNA and restore CFTR function (Oren et al. Journal of Cystic Fibrosis 2021). We are currently expanding our personalized medicine research, implementing the ASO approach in patient-derived intestinal organoids which serve as biomarkers predicting clinical benefit from CFTR modulator therapies.

A scheme of the splicing pattern generated from the 3849+10 kb C-to-T splicing mutation and its effect on the inclusion of the 84 bp cryptic exon in the mature mRNA. The cryptic exon contains an in-frame stop codon, which leads to the production of reduced levels of CFTR transcripts, due to degradation by the NMD mechanism, and truncated nonfunctional CFTR protein.

Intestinal organoids, derived from a CF patient carrying a splicing mutation, treated with an ASO designed to mask specific splicing motifs and restore correct splicing pattern.
- Nonsense Mutations
-
10-15% of CF-causing mutations are nonsense mutations which lead to premature termination codon (PTC). Transcripts carrying PTCs undergo degradation by the Nonsense Mediated mRNA Decay (NMD) pathway. Various small molecules, which can read-through PTCs, permit translation of full-length proteins. However, this approach is dependent on NMD efficiency and thus may limit the response of patients to the treatment. Using various cellular models, among them patient-derived epithelial cells, we are investigating a novel therapeutic strategy aimed to stabilize NMD-prone CFTR transcripts by epigenetic modulation of m6A RNA methylation (in collaboration with Prof. Rotem Karni).

Methylation of Adenine on position 6 in RNA molecules (m6A) plays a key role in RNA biogenesis. M6A marks the translation termination codon and helps the NMD machinery to distinguish it from premature termination codons (PTCs). It was recently discovered that m6A modulation can inhibit NMD and stabilize unstable mRNA molecules carrying PTCs.

